Prof. Dror Noy : Bioenergy and Protein Design
Research Overview
Our goal is to learn how Nature exploits fundamental physical principles in the assembly and operation of light energy conversion systems, and to find ways to implement the same strategies in man-made solar fuels production systems by integrating natural and artificial components. The main research effort in our lab focuses on constructing simple and robust functional analogs of the natural photosynthetic light-harvesting complexes and reaction centers. Designing analogs of light-harvesting complexes requires proteins that self-assemble and organize natural or modified pigments in dense and highly absorbing arrays. Designing reaction centers analogs requires the assembly of pigments and redox cofactors in electron transport chains for stable photoinduced charge separation. The benefits of using designed protein-pigment complexes are the ability to determine both structure and function by amino acid sequences, the availability of recombinant DNA techniques for preparing large quantities of proteins, and the straightforward interfaces to natural components that promises smooth integration with natural systems. Thus, in parallel to our main effort, we are trying to construct systems for “photosynthesis outside the membrane” that will enable integration of natural photosynthetic and artificial components into solar energy conversion devices outside the native biological context.
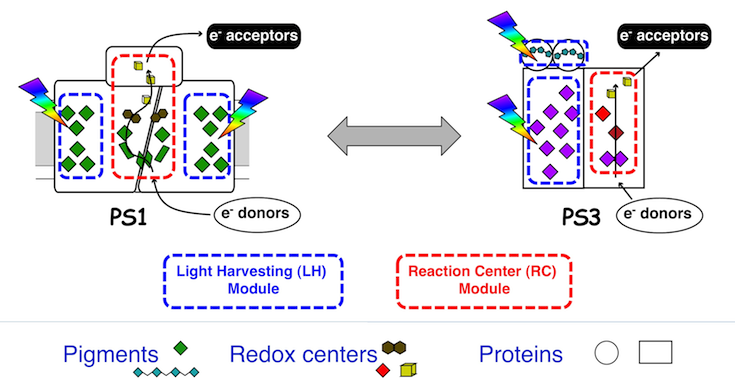
Design of an artificial solar energy converter by reverse engineering natural photosystems. Our ERC-funded project aims at designing and constructing an artificial solar energy converter that is inspired by, but does not necessarily mimic, the fundamental solar energy conversion unit of natural photosynthesis – the photosystem. The new system, code named PS3 will be designed as a water-soluble functional analog of the natural photosystems, PS1 and PS2.
Protein Design
Many medium- to high-resolution structures of natural redox enzymes and photosynthetic protein complexes are a source of inspiration and an excellent starting point for new protein design. We collaborate with leading experts of protein computational design in order to implement our insights from the natural structures in state-of-the-art computational tools such as the protCAD, and Rosetta software packages. In addition to identifying and implementing general features of natural cofactor-binding sites in de novo designed protein scaffolds, we use computational tools for taking cofactor-binding structural motifs out of their natural context, and adapting these to fold as small independent proteins.
The output of the design stage is amino acid sequences of new proteins that are coded into DNA and over-expressed in E. coli bacteria. Since proteins are designed to spontaneously assemble with cofactors, preparing the holoprotein usually requires simply mixing the apoprotein with the desired cofactors. In some cases we need to assemble hydrophobic water-insoluble native chlorophylls with water-soluble proteins; for these we are developing new assembly methods based on water-in-oil emulsions. New designs are tested by a variety of analytical and spectroscopic methods, and the results are used for optimizing the previous designs in an iterative process.
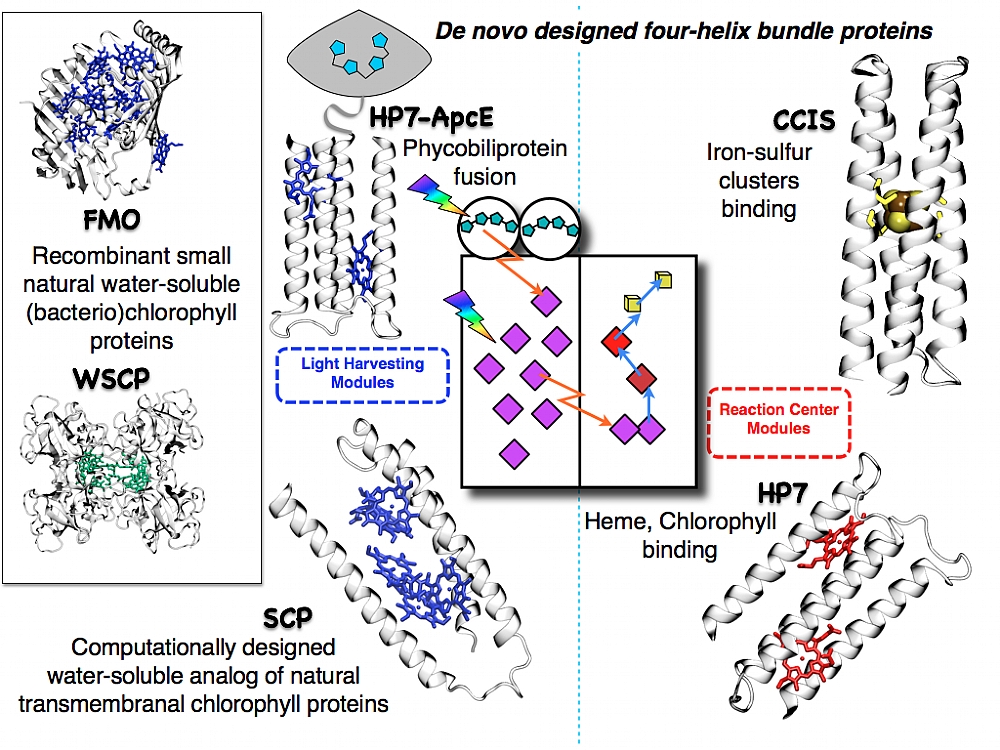
Protein-cofactor building blocks for an artificial water soluble photosystem. A variety of proteins ranging from completely de novo designed to variants of native proteins are developed as functional analogs of the natural lighte harvesting complexes and reaction centers. These will eventually be integrated into a functional photosystem.
Research Projects
- De novo designed four-helix bundle proteins
- binding water-soluble bacteriochlorophyll derivatives
- binding iron-sulfur clusters
- Computational design of a water-soluble analog of natural transmembranal chlorophyll-binding proteins
- Chlorophyll-binding protein/phycobiliprotein fusion
- Water-in-oil emulsions for self-assembly and screening of water-soluble protein complexes with hydrophobic chlorophyll derivatives
Photosynthesis Outside the Membrane
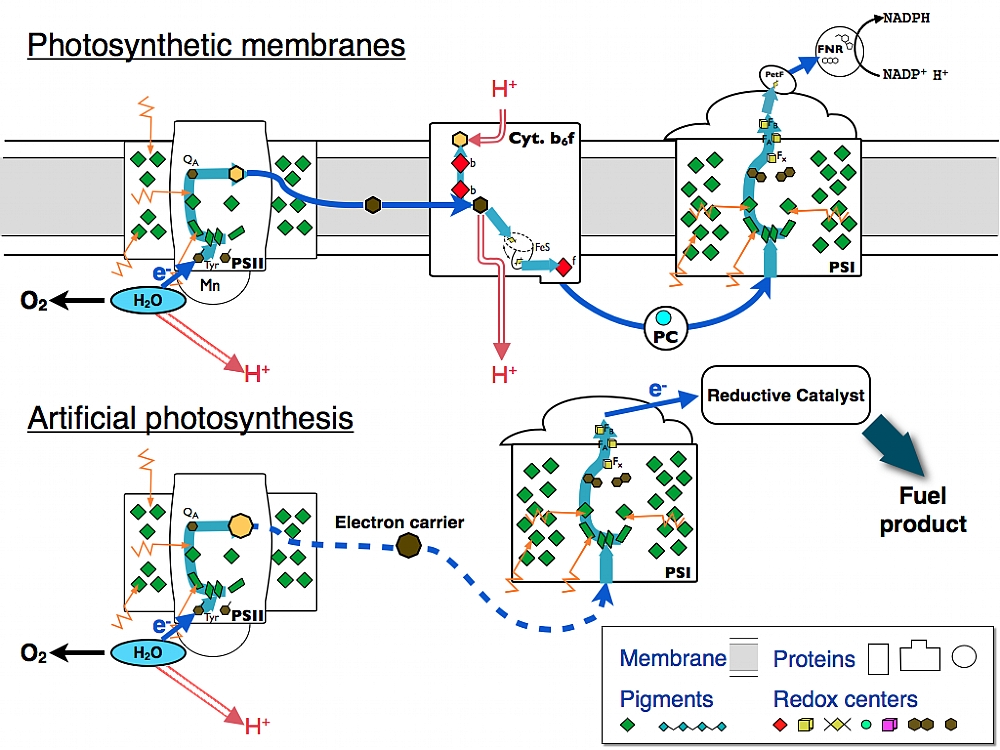
Early in the course of evolution, living organisms have committed to ATP as the universal energy carrier, and to energy transducing membranes and proton motive force (PMF) for driving ATP production. Since human technology relies on electron motive force (EMF), this commitment has made adapting biological energy conversion systems to technological applications a very difficult challenge. Yet, natural energy converters offer unique advantages over artificial solar energy converters, particularly in providing elaborate and efficient catalytic modules for driving difficult redox reactions such as water oxidation, and hydrogen production. In order to make use of these catalysts in a non-biological setting, we are trying to develop hybrid natural/artificial systems where natural photosynthetic components operate outside the membrane and are coupled to redox catalysts that are not necessarily part of the native photosynthetic energy conversion process.
Taking natural photosystems out of the biological context. The natural photosystems can be used for driving redox reactions in water. We are working to couple photosystem II to photosystem I, and photosystem I to reductive catalysts in order to drive electrons all the way from water to reduction of small organic and inorganic molecules that may serve as alternative fuels.
Research Projects
- Microencapsulation and coupling of natural photosystems in sol-gel matrices
- Re-engineering the electron transport chain of photosystem I for driving hydrogen photoproduction
- PEPDIODE – Peptide based diodes for solar cells
